


THE MULTICOLOR FLUORESCENCE IN SITU
HYBRIDIZATION (mFISH) HOMEPAGE
Liehr T*
*Corresponding Author: Dr. Thomas Liehr, Institut für Humangenetik und Anthropologie,
Kollegiengasse 10, D-07743 Jena, Germany; Tel.: +49-3641-935533; Fax: +49-3641-935582;E-mail: i8lith@mti.uni-jena.de
page: 27
|
|
CONTENT
I have collected all the literature on mFISH and mFISH applications, and made the data available on the mFISH Homepage [11], which was created and has been available online since 2002. All literature that applies mFISH approaches according to our aforementioned definition is collected from publications available to the author and from PubMed (http:// www.ncbi.nlm.nih.gov/sites/ entrez) and is arranged according to the scheme shown in Figure 1. There are nine main categories: 1) Introduction gives basic information on the page; 2) Reviews lists papers that provide an overview on mFISH techniques and their pitfalls; 3) mFISH using wcps is subdivided into literature on M-FISH (multiplex-FISH)/ SKY (spectral karyotyping) applied in human Clinical Cytogenetics, Tumor Cytogenetics and Others; 4) Fish-banding summarizes all available approaches using human probes to achieve an mFISH-based chromosome banding; 5) CEP & SC-probes reviews all papers on mFISH that use human centromeric and single copy probes; 6) Additional mFISH contains human mFISH probe sets not fitting any previous category; 7) mFISH in animal and 8) mFISH in plant are put together under these buttons. The corre sponding subchapters can be seen in Figure 1. Under 9) mFISH probes available in cooperation and/or at cost price from the author of this article are listed. The mFISH Homepage [11] can be used as a source of information in many ways. It can provide an overview into the importance of the mFISH field itself by taking note of the sheer volume of studies that apply multicolor molecular cytogenetics. It facilitates the search for mFISH reviews and basics. Also, it provides all relevant literature on already performed mFISH studies in clinical syn dromes, special malignancies, mutagenesis, radiobiology, linkage/mapping, interphase-architecture and Zoo- FISH (Figure 2).It is always up-to-date on technical developments and new probe sets [11]. It provides information on mFISH combined with immunohistochemistry, microdissection after mFISH, and alternatives to mFISH approaches. For FISH-banding [13], the following probe sets can be used: arm specific probes, M-FISH combined with banding, the chromosome bar code, Rx-FISH (cross-species color banding), spectral color banding (SCAN) and multicolor banding (MCB)/m-banding (Figure 3). Single copy probes and/or centromeric probes are applied in many different combinations and various applications; mFISH sets using sub-telomeric or microdeletion syndrome specific probes (“MultiFISH” assay) were created as well as for gene mapping, prenatal, preimplantation and clinical diagnostics 30 or tumor cytogenetics. Furthermore, mFISH using alphoid probes, was developed for different applications, e.g., characterization of small supernumerary marker chromosomes [14]. While the aforementioned pages of the Homepage are mainly concentrated on human cytogenetics, mFISH applied in animals and plants are also included. Besides ~60 papers on mFISH using murine probes, also included are mFISH probe sets for Nasonia (Pteromalidae: Hymen optera), chicken, rat, dog and horse (see FISH-banding) were reported in at least one publication, each. For plants only one mFISH application has been reported in Acri cotopus (Figure 4). In summary, the mFISH Homepage is a valuable resource for all working in the diagnostic and research field of molecular cytogenetics.
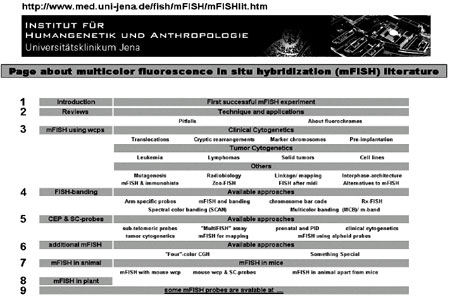
Figure 1. Interactive start page of the regularly updated mFISH Homepage. The figures 1-9 are not visible on the web page.
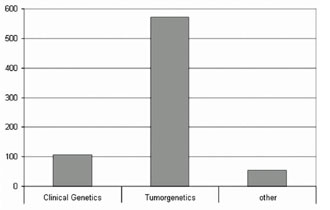
Figure 2. Number of published papers applying M-FISH/SKY in clinical genetics,tumor cytogenetics and other fields. Up to 04/2008 >700 publications used this mFISH approach, i.e., FISH using all 24 human whole chromosome painting libraries as probes, simultaneously.
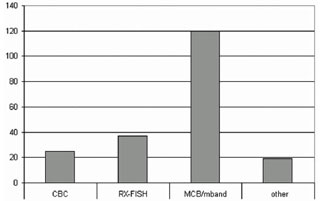
Figure 3. The FISH-banding approaches have been used in >200 research studies. The chromosome bar code (CBC), Rx-FISH and multicolor banding (MCB/m-band) are there most frequently applied.
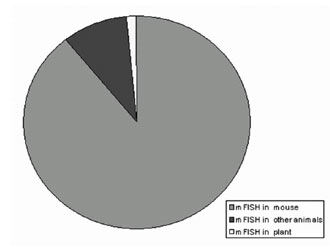
Figure 4. In animal and plants, mFISH is mainly applied in the mouse (Mus musculus).
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

