


MOLECULAR ANALYSIS OF SURVIVAL MOTOR
NEURON AND NEURONAL APOPTOSIS INHIBITORY
PROTEIN GENES IN MACEDONIAN SPINAL
MUSCULAR ATROPHY PATIENTS
Kocheva SA1,2, Vlaski-Jekic S3, Kuturec M2, Efremov GD1,*
*Corresponding Author: *Corresponding Author: Professor Dr. Georgi D. Efremov, Macedonian Academy of Sciences
and Arts, Research Centre for Genetic Engineering and Biotechnology, Aven Krste Misirkov 2,
POB 428, 1000 Skopje, Republic of Macedonia; Tel: +389-2-3235411; Fax: +389-2-3115434;
E-mail: gde@manu.edu
page: 55
|
|
MATERIALS AND METHODS
Patients. A total of 30 unrelated SMA patients (17 with type I, eight with type II and five with type III form of the disease), 30 parents and 30 unrelated healthy individ uals, were studied. Patients fulfilled the diagnostic criteria defined by the International SMA Consortium [21].
Molecular Analysis. Genomic DNA was extracted from peripheral blood lymphocytes following standard phenol/chlorophorm procedures [22]. The presence of deletions of exons 7 and 8 of the SMN gene were deter mined from polymerase chain reaction/restriction fragment length polymorphism (PCR/RFLP) analyses [23]. Poly merase chain reaction was carried out in a final volume of 25 mL containing 100 ng of genomic DNA, 20 pmoles of each primer, 200 mM of each dNTP, 1.5 mM MgCl2 and 1U Taq DNA polymerase (Ampli Taq Gold; Applied Bio Systems, Branchburg, NJ, USA). Sequences of the primers for amplification of SMN exon 7 (R111: 5’-AGA CTA TCA AAC TTA AAT TTC TGA TCA-3’ and x-7Dra: 5’-CCT TCC TTC TTT TTG ATT TTG TTT-3’) and SMN exon 8 (541C960: 5’-GTA ATA ACC AAA TGC AAT GTG AA-3’ and 541C1120: 5’-CTA CAA CAC CCT TCT CAC AG-3’) [23]. Samples were subjected to 30 cycles of amplification consisting of 1 min. at 94°C, 1 min. at 57°C and 1 min. at 72°C and a final extension at 72°C for 10 min. The PCR products of SMN exon 7 and SMN exon 8 were digested with 2U of restriction enzymes DraI and DdeI, respectively, at 37°C overnight. Digested products were run on 8% non denaturing acrylamide gel (39:1 Acryla mide/BisAcrylamide) at 30mA for 3 hours in TBE buffer and visualized under UV light after ethidium bro mide staining. The PCR digestion products of exons 7 and 8 of the SMNtel gene and the copy gene can readily be distinguished since the copy gene contains a recognition site for the restriction enzyme (DraI/exon 7 and DdeI/exon 8) and that site is absent in the SMNtel (functional) gene. The SMN/exon 7/DraI digestion in healthy individuals, converged into two fragments to the SMNtel and SMNcen copy. The SMN/exon 8/DraI digestion gave three frag ments: that with a higher molecular weight corresponding to SMNtel, and the rest corresponding to the SMNcen copy. Normal results were indicated by the presence of SMNtel and SMNcen exons 7 and 8 (Figure 1A and 1B). The restriction digest method does not differentiate between a true deletion and a gene conversion, but cor rectly determines the absence of SMNtel exons 7 and 8 from their normal genomic position.

Figure 1. A) The PCR/RFLP analysis of exon 7. Lanes 1and 2: patients with homozygous deletion of SMNtel exon 7; lanes 3-7: normal pattern. B) The PCR/RFLP analysis of exon 8. Lane 1: undigested sample; lanes 2 and 3: patients with
homozygous deletion of SMNtel exon 8; lanes 4-6: normal pattern.
Single-Strand Conformation Polymorphism Analy ses of Survival Motor Neuron Exons 7 and 8. Single-strand conformation polymorphism (SSCP) analyses were carried out as described in (9). Genomic DNA (100-200 ng) was amplified using exon 7 R111 and 541C770 and exon 8 541C960 and 541C1120 specific oligonucleotide primers. A total of 10 mL of PCR product was mixed with 10 mL 95% formamide, 0, 25% bromphenol blue and 0.25% xylene cyanol. The samples were denatured at 95°C for 5 min. and loaded onto a 12% non denaturing acryl amide gel (39:1 Acrylamide/BisAcrylamide). The gels were run at 15W for 16 hours at 16°C and visualized under UV light or after silver staining (Figure 2). The results were compared with those from DNA from healthy con trols and from individuals with deletions confirmed by the restriction method.
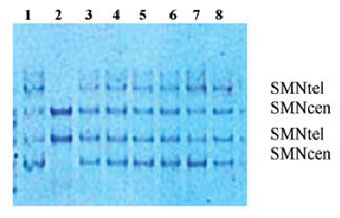
Figure 2. The SSCP analysis of exon 7 of the SMN gene. Lane 1: normal control; lane 2: patient with homozygous deletion of SMNtel copy; lanes 3-8: normal pattern.
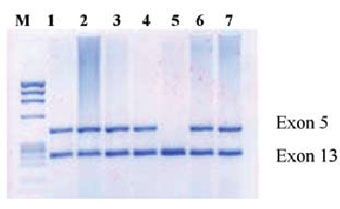
Figure 3. The PCR analysis of the NAIP gene. Lane 1: normal control; lanes 2-4, 6 and 7: normal pattern; lane 5: homozygous deletion of exon 5.
Analysis of the Neuronal Apoptosis Inhibitory Pro tein Gene. The NAIP gene analysis was performed by multiplex PCR amplification of exons 5 and 13, using primers specific for exon 5 (1863: 5’-CTC TCA GCC TGC TCT TCA GAT-3’ and 1864: 5’-AAA GCC TCT GAC GAG AGG ATC-3’) and exon 13 (1258: 5’-ATG CTT GGA TCT CTA GAA TGG-3’ and 1343: 5’-CCA GCT CCT AGA GAA AGA AGG A-3’) as described in [7]. Exon 13 is present in both functional and pseu-dogene copies of NAIP and can therefore be used as a positive PCR control for exon 5 which is present only in the func tional NAIP gene. The PCR was performed using the pro cedure described above, except that the annealing step was carried out for 1 min. at 60°C. Samples were loaded onto a 2% agarose gel containing ethidium bromide, run for 1 hour at 80-100V and visualized under UV light (Figure 3).
Table 1. Deletion genotypes including SMN (exon 7 and 8) and NAIP (exon 5) genes
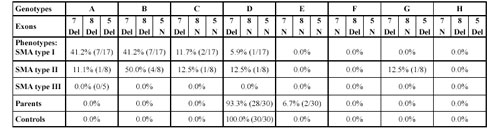
The pattern includes SMNtel exon 7, SMNtel exon 8, and NAIP exon 5. Del: homozygous deletion; N: without a homozygous deletion.
Analyses of Exons 7 and 8 of the Survival Motor Neuron Gene. Table 1 summarize the results of the screen ing of 30 SMA individuals. Seventy-seven percent of all the SMA patients (94.0% of type I and 87.5% of type II) lacked the telomeric copy of the SMN gene. Of these, 83.0% (19/23) had deletions of both exons and 17.4% (4/23) had deletion of only exon 7. The deletion was most common in patients with type I of the disease (94.1%), less common in patients with type II (87.5%). We did not detect a deletion of exons 7 and/or 8 in patients with type III. The centromeric copies of exons 7 and 8 were not affected in any of the patients, however, a deletion of one copy was found in one normal control. The SSCP analysis detected an aberrant fragment indicating a point mutation in exon 7 of the SMNtel gene in one type I patient.
Analyses of the Neuronal Apoptosis Inhibitory Protein Gene. Deletion of exon 5 of the NAIP gene was found in 41.2% (7/17) type I patients and in one type II patient (1/8). Only two of 30 parents (6.7%) were found to be homozygous for the NAIP exon 5 deletion. No dele tions of the NAIP exon 5 were found in 30 normal con trols.
Correlation Analyses of Survival Motor Neuron and Neuronal Apoptosis Inhibitory Protein Gene Dele tions. In order to correlate the extent of the deletion with clinical severity of disease, we have analyzed the deletion pattern of both genes. We have constructed genotypes of patients using the model proposed in [24]. The results are shown in Table 1. Genotype A, which represents a large deletion, including telomeric SMN exons 7 and 8 and also NAIP, has a higher incidence in type I SMA patients (41.0%) compared with type II (11.1%). Genotype B, which represents smaller deletions (deletions of telomeric SMA exons 7 and 8), is predominantly found in type II SMA (50.0%). Genotype C (deletion of exon 7) was found in three patients (two with type I and one with type II) and genotype D (no deletions of SMNtel exons 7 and 8 and NAIP) in one patient with type I, one with type II and five patients with type III. Genotype E, which represents dele tions only in NAIP, was found in 2/30 (6.7%) carriers. Genotype G, which represents deletions in exon 7 of SMN and exon 5 of NAIP, was found in one patient with type II SMA. Deletion only in SMNtel exon 8 (genotype F) and in SMNtel exon 8 and exon 5 of NAIP (genotype H) was not found in our samples.
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

