


UNIPARENTAL DISOMY (UPD): A CONSEQUENCE
OF NON-DISJUNCTION AND THE IMPLICATIONS
IN PRENATAL DIAGNOSIS
Velissariou V*
*Corresponding Author: Dr. Voula Velissariou, Cytogenetics Laboratory, Department of Genetics and Molecular Biology, Mitera Hospital, Erythrou Stavrou 7, Maroussi, Athens 11523, Greece; Tel.: +30-210-686-9869; Fax: +30-210-689-9476; E-mail: voulavel@ hol.gr
page: 55
|
|
UNIPARENTAL DISOMY STUDIES IN PRENATAL DIAGNOSIS
According to the recent literature, UPD molecular studies should be performed in the following cases during prenatal diagnosis:
a) When mosaicism for chromosomes 6, 7, 11, 14, 15 and 16 is detected after chorionic villus sampling (CVS) and subsequent amniocentesis shows a normal fetal karyotype, most likely caused by “trisomy rescue”. Theoretically, in one-third of these cases UPD should be present in the euploid cell lines. For chromosome 20, evidence has accumulated during the past few years that it may be responsible for abnormalities, when both chromosomes 20 are maternally derived, and it is advisable to offer UPD analysis [11,13,14]. For chromosomes 2, 4 and 5, there is still no convincing evidence that UPD may cause abnormalities in order to justify molecular studies, when detected in a mosaic state in the placenta [5].
b) When a non homologous familial or de novo Robertsonian translocation involving chromosomes 14 and 15, known to contain imprinted genes, is detected either in CVS or amniotic fluid cells. According to Silverstein et al. [7], the risk estimate is 0.65%, which justifies UPD analysis in fetuses diagnosed prenatally with Roberstonian translocations. Based on this risk the same authors suggest testing these fetuses for UPD in amniocytes rather than chorionic villi, when the risk for unbalanced karyotype is ~1%, comparable to the risk for UPD.
c) When a homologous de novo Robertsonian translocation or an isochromosome involving either chromosome 14 or 15 is detected. Berend et al. [15] estimated that the risk of UPD, when a homologous acrocentric rearrangement is identified prenatally, is approximately 66%, which certainly justifies UPD analysis.
d) When a 47,+ESAC (extra structurally abnormal chromosome) karyotype is detected, it may infrequently coexist with UPD for the same chromosome that the ESAC was derived from; it may be that the zygote from a 23,abn + 23,N (46,abn) conception attempted to correct the imbalance, or at least lessen it, by replication of the normal homologue [16]. If the ESAC is derived from chromosome 15, known to contain imprinted genes, it is advisable to perform UPD analysis.
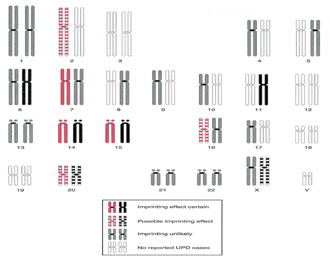
Figure 2. Map of imprinted regions in the human genome, based on phenotypes detected in cases of UPD involving the maternally inherited homologues (left chromosome in each pair) or the paternally inherited homologues (right chromosome in each pair).[Modified from Ledbetter DH, Engel E: Uniparental disomy in humans: Development of an imprinting map and its implications for prenatal diagnosis. Hum Mol Genet 1995; 4: 1757-1764.]
|
Chromosome UPD and Inheritance |
Associated Genetic Disease or Abnormalities |
|
Paternal UPD 6 |
Transient neonatal diabetes mellitus |
|
Maternal UPD 7 |
Silver-Russell syndrome |
|
Paternal UPD 11 |
Beckwith-Wiedemann syndrome |
|
Maternal UPD 14 |
Hypotonia, motor development delay, mild dysmorphic facial features, low birth weight, growth abnormalities |
|
Paternal UPD 14 |
Severe mental and muscoskeletal abnormalities |
|
Maternal UPD 15 |
Prader-Willi syndrome |
|
Paternal UPD 15 |
Angelman syndrome |
|
Maternal UPD 16 |
Intrauterine growth retardation |
|
Maternal UPD 20 |
Intrauterine growth retardation and/or postnatal growth retardation |
Table 2. Uniparental disomy and associated genetic disease.
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

