


CHROMOSOMAL MOSAICISM IN THE PATIENTS WITH TURNER'S SYNDROME
Vyatkina S1., Nagornaya I1,3., Loginova J1,3., Lyazina L2., Vasileva I2., Prozorova M2., Verlinskaya D2., Kuznetzova T 1 Baranov V.S. 1
*Corresponding Author: Professor Dr. Vladislav S. Baranov, Laboratory of Prenatal Diagnosis of Inherited Disorders, 1The D. O. Ott Institute of Obstetrics & Gynecology, the Russian Academy of Medical Sciences, Mendeleevskaya l., 3, Saint Petersburg, Russia fax +7 (812) 328 04 87; e-mail: svetavyach@mail.ru
2 Saint Petersburg Centre for Medical Genetics, Tobolskaya ul., 5, Saint Petersburg, Russia
fax +7 (812) 542 67 76, e-mail: gkdmgenc@zdrav.spb.ru
3Russian-Finnish Medical Centre "AVA-Peter", Nevskii pr., 22/24, Saint Petersburg, Russia
fax: +7 (812) 314 51 19, e-mail: avapeter@bcltele.com
page: 17
|
|
RESULTS AND DISCUSSION
Karyotyping of PHA-stimulated lymphocytes has proved 45,X karyotype in 41 of total 88 patients studied, karyotype 46,XX - in 1 case and 46,X,der (X) - in 4 cases. Mosaic karyotypes with admixture of 45 cell lines with normal or abnormal ?-, Y- or marker chromosomes were revealed in 42 cases. FISH analysis with X-specific DNA-probes proved the existance of additional cell lines with aberrant X-chromosomes in 5 out of 41 patients with 45,X. PCR analysis of DNA samples from 34 patients with monosomy X revealed the presence of Y specific DNA sequences in 2 cases.
Taking into account that the rate of chromosomal mosaicism depends on the number of the scored cells, on the methods applied (Hook, 1977; Binder et al., 1995), as well as on the type of tissues we tried to study the cells of different germ-line origin.
The combined FISH analysis of lymphocytes and buccal mucosal cells was achieved in 69 cases. Karyotype 45,X was ascertained in 29 cases, karyotype 46, X,der(X) in both tissues studied was recorded in one case. In 4 patients (10%) mosaic karyotypes (45,X/46X,der(X)) has been proven in peripheral blood lymphocytes in one case and in buccal mucosa cells in 3 cases (tab. 1, ? 39 and ?? 1, 11, 22 accordingly). Chromosomal mosaicism in both tissues was revealed in 35 from 39 patients (90 %).
Obvious quantitative differences of mosaicism in cell lines of different tissues have been detected in overwhelming majority of mosaics (tab. 1). The preponderence of one cell line in two tissues was found in 28 out of 35 cases (80%) studied. In 7 patients the cell clone, minor in one tissue, was dominant in other tissue (?? 4, 6, 9, 10, 20, 28, 33, tab. 1). The portion of cells with aberrant X-chromosome in "cryptic" mosaics was 5-8 % in blood and 12-22 % - in buccal mucosa (?? 1, 15, 17, 23, 25, tab. 1).
Cryptic mosaicism involving Y-chromosome was found in 6 % and X-chromosome – in 15% patients with a pure X monosomy detected by standard cytogenetic analysis. Karyotype 45, X in both tissues has been assessed in the majority of patients. According to availble data the frequency of "cryptic" mosaicism varies from 0 % to 61 % for Y-chromosome (Lorda-Sanchez et al., 1992; Coto et al., 1995; Yorifuji et al., 1997) and from 2,4 % to 90 % for X-chromosome (Yorifuji et al., 1997; Nazarenko et al., 1999; Fernandez-Garcia et al., 2000).
Substantial fluctuation in the rate of mosaicism could be attributed to the sample’s volume as well as to the sensitivity of the techniques applied. Owing to its high sensitivity and specificity, PCR technique readily reveals chromosomal mosaicism attributed to extra genetic material but it does not permit estimation of a cell line ratio. The quantitative characteristic of cell lines in mosaics might be achieved by means of FISH analysis of interphase nuclei. Meanwhile spontaneous aneuloidy and hybridization efficiency might substantially influence the results. 5% cut-off value for mosaicism both in peripheral blood, and in buccal mucosa was chosen and this value roughly corresponded to the background level of spontaneous sex chromosomes aneuploidy (Eastmond, Pinkel, 1991). However, quantitative variations of X-chromosome aneuploidy in different tissues can vary (2 %-4,04 % for non-cultivated and cultivated lymphocytes respectively and 1,79 % for native buccal mucosal cells) (Nazarenko, etc., 1997; Jacobs et al., 1997). It should be menthioned that the number of marker loci detected by specific DNA probes might remain constant in spite of structural chromosomal abnormalities and the relevent cell lines with abnormal karyotype could not be detected by FISH analysis. Thus the results of FISH analysis in interphase nuclei could not precisely correspond to metaphase studies.
Table 1. Molecular-cytogenetic characteristic of mosaic karyotypes
# |
Lab. index |
blood lymphocytes (metaphase plate n=100) |
blood lymphocytes (interphase nucleus n=500) |
buccal mucosal cells
(interphase nucleus n=500) |
1 |
760 |
45,? |
DXZ1?1[482], DXZ1?2[18] |
DXZ1?1[438], DXZ1?2[61] |
2 |
93 |
45,?[88]/47,???[5]/46,??[7] |
DXZ1?1[409], DXZ1?3[44], DXZ1?2[47] |
DXZ1?1[375], DXZ1?3[44], DXZ1?2[81] |
3 |
15 |
45,?[91]/46,??[9] |
DXZ1?1[471], DXZ1?2[29] |
DXZ1?1[427], DXZ1?2[73] |
4 |
319 |
45,?[40]/46,??[60] |
DXZ1?1[248], DXZ1?2[252] |
DXZ1?1[376], DXZ1?2[124] |
5 |
419 |
45,X[71]/46,XX[29] |
DXZ1?1[328], DXZ1?2[172] |
DXZ1?1[349], DXZ1?2[151] |
6 |
632 |
45,X[41]/46,XX[59] |
DXZ1?1[222], DXZ1?2[278] |
DXZ1?1[266], DXZ1?2[234] |
7 |
422 |
45,X[88]/46,XX[12] |
DXZ1?1[449], DXZ1?2[51] |
DXZ1?1[413], DXZ1?2[87] |
8 |
1161 |
45,X[8]/46,XX[92] |
DXZ1?1[33], DXZ1?2[467] |
DXZ1?1[40], DXZ1?2[460] |
9 |
1323 |
45,X[61]/46,XX[39] |
DXZ1?1[321], DXZ1?2[179] |
DXZ1?1[115], DXZ1?2[385] |
10 |
1062 |
45,?[89]/47,???[6]/46,??[5] |
DXZ1?1[426], DXZ1?3[42], DXZ1?2[32] |
DXZ1?3[443], DXZ1?1[23], DXZ1?2[34] |
11 |
874 |
46,?? |
DXZ1?2 |
DXZ1?1[72], DXZ1?2[428] |
12 |
43 |
45,?[85]/46,?,r(?)[15] |
DXZ1?1[442], DXZ1?2[58] |
DXZ1?1[400], DXZ1?2[100] |
13 |
28 |
46,?,r(?)[67]/45,?[33] |
DXZ1?2[369], DXZ1?1[131] |
DXZ1?2[363], DXZ1?1[137] |
14 |
4 |
46,?,r(?)[64]/45,?[36] |
DXZ1?2[344], DXZ1?1[156] |
DXZ1?1[362], DXZ1?2[138] |
15 |
82 |
45,X[92]/46,X,r(X)[8] |
DXZ1?1[471], DXZ1?2[29] |
DXZ1?1[395], DXZ1?2[105] |
16 |
107 |
45,X[87]/46,X,r(X)[13] |
DXZ1?1[411], DXZ1?2[89] |
DXZ1?2[267], DXZ1?1[233] |
17 |
194 |
45,X[95]/46,X,r(X)[5] |
DXZ1?1[474], DXZ1?2[26] |
DXZ1?1[414], DXZ1?2[86] |
18 |
1529 |
45,X[60]/46,X,r(X)[35]/47,X,r(X), +r(X)[5] |
DXZ1?1[321], DXZ1?2[154], DXZ1?3[25] |
DXZ1?2[304], DXZ1?1[196] |
19 |
33 |
45,?[80]/46,?,r(?)[10]/46,?,idic(X)(q22)[5]/47,?,r(?),+r(?)[5] |
DXZ1?1[390], DXZ1?2[48], DXZ1?3[62] |
DXZ1?1[428], DXZ1?2[72] |
20 |
680 |
46,X,der(X)[51]/46,X,r(X)[29]/45,X [15]/46,X,dicr(X)[5] |
DXZ1?2[405], DXZ1?1[69], DXZ1?3[26] |
DXZ1?1[285], DXZ1?2[189], DXZ1?3[26] |
21 |
220 |
45,?[81]/46,X,i(X)(q10)[19] |
DXZ1?1[401], DXZ1?2[99] |
DXZ1?1[405], DXZ1?2[95] |
22 |
914 |
46,X,i(X)(q10) |
DXZ1?2[490], DXZ1?1[10], |
DXZ1?2[473], DXZ1?1[27], |
23 |
85 |
45,X[92]/46,X,idic(X)(p11)[8] |
DXZ1?1[466], DXZ1?3[34] |
(DXZ1?1[417], DXZ1?3[83] |
24 |
901 |
45,X[67]/46,X,idic(X)(p11)[33] |
DXZ1?1[335], DXZ1?3[165] |
DXZ1?1[337], DXZ1?3[163] |
25 |
558 |
45,?[95]/46,X,idic(X)(q22)[5] |
DXZ1?1[469], DXZ1?3[31] |
DXZ1?1[392], DXZ1?3[108] |
26 |
581 |
45,X[70]/46,X,dup(X)(p21p22)[25]/46,XX[5] |
|
|
27 |
151 |
45,X[79]/46,X,der(X)[21] |
DXZ1?1[403], DXZ1?2[97] |
DXZ1?1[326], DXZ1?2[174] |
28 |
313 |
46,X,der(X)[62]/45,?[33]/46,X, dicder(X)[5] |
DXZ1?2[318], DXZ1?1[154], DXZ1?3[28] |
DXZ1?1[467], DXZ1?2[33] |
29 |
1442 |
46,X,der(X)[53]/45,?[42]/47,X,der (X),+der(X)[5] |
DXZ1?2[269], DXZ1?1[204], DXZ1?3[27] |
DXZ1?2[295], DXZ1?1[205] |
30 |
166 |
46,X,del(X)(qter-p11.2:)[95]/45,?[5] |
DXZ1?2[469], DXZ1?1[31] |
DXZ1?2[430], DXZ1?1[70] |
31 |
880 |
45,X[54]/46,X,del(X)(qter-p11.2:) [46] |
DXZ1?1[253], DXZ1?2[247] |
DXZ1?1[374], DXZ1?2[126] |
32 |
156 |
46,?,del(X)(pter-q23:)[77]/45,X[23] |
DXZ1?2[391], DXZ1?1[109] |
DXZ1?2[434], DXZ1?1[66] |
33 |
1314 |
45,?[57]/46,?Y[43] |
DYZ3?0[290], DYZ3?1[210] |
DYZ3?1[282], DYZ3?0[218] |
34 |
748 |
47,XYY[28]/45,X[14]/46,XY[58] |
DYZ3?2[144], DYZ3?0[63], DYZ3?1[290] |
DYZ3?2[98], DYZ3?0[183], DYZ3?3[25], DYZ3?1[194] |
35 |
654 |
46,?,dic(Y)[86]/45,?[14] |
DYZ3?2[441], DYZ3?0[59] |
DYZ3?2[303], DYZ3?0[197] |
36 |
19 |
46,?,r(Y)[7]/45,?[6]/46,?,dicr(Y) [5]/46,?Y[82] |
DYZ3?1[442], DYZ3?0[27], DYZ3?0[31] |
DYZ3?1[328], DYZ3?0[172] |
37 |
1065 |
46,X,dicder(Y)[66]/45,X[34] |
DYZ3?2[333], DYZ3?0 [167] |
DYZ3?2[305], DYZ3?0 [173], DYZ3?4[22] |
38 |
1445 |
45,X[70]/46,X,dicder(Y)[30] |
DYZ3?0[362], DYZ3?2[138] |
DYZ3?0[402], DYZ3?2[98] |
39 |
1511 |
45,X[73]/46,X,der(Y)[27] |
DYZ3?0[371], DYZ3?1[129] |
DYZ3?0 |
Meanwhile, limited tissue mosaicism (tab. 2) revealed by different methods could be proved in only about 20 % of cases. Therefore, the hypothesis that the patients possessing 45,X karyotype in their lymphocytes, should have additional cell lines with normal or aberrant sex chromosomes (Hook, Warburton, 1983) is fair only for a part of patients. True chromosomal mosaicism in all tissues, irrespective of their germinal origin is typical for the most of TS cases. Domination of the same cell line in the tissues of different embryonic origin is encountered in more than 70 % cases
Table 2. Cell lines distribution in different tissues of Turners syndrome patients .
Authors |
Number of cases |
Analyzed
tissue |
Research method |
Character of mosaicism |
Character of cell line domination |
True |
limited |
In two tissues |
In one tissue |
Mashkova M.V., 1974 |
3 |
lymphocytes, skin fibroblasts |
cytogenetic |
3 |
0 |
2 |
1 |
Held et al., 1992 |
56 |
lymphocytes, skin fibroblasts |
cytogenetic |
32 |
24 |
24 |
8 |
Reddy, Sulcova, 1998 |
5 |
lymphocytes, ovarian tissue |
fish |
5 |
0 |
4 |
1 |
Nazarenko et al.,1999 |
13 |
lymphocytes, buccal mucosal cells |
fish |
9 |
4 |
2 |
7 |
Hanson et al., 2001 |
35 |
lymphocytes, buccal mucosal cells |
fish |
32 |
3 |
- |
- |
Hanson et al., 2002 |
11 |
lymphocytes, buccal mucosal cells |
fish |
11 |
0 |
8 |
3 |
Fernandez et al., 2000 |
2 |
lymphocytes, ovarian tissue |
fish |
2 |
0 |
- |
- |
Osipova et al., 1998 |
2 |
leukocytes, ovarian tissue |
PCR |
1 |
1 |
- |
- |
Mendes et al., 1999 |
2 |
leukocytes, ovarian tissue |
PCR |
2 |
0 |
- |
- |
Own data |
39 |
lymphocytes,
buccal mucosal cells |
fish |
35 |
4 |
28 |
7 |
summary |
168 |
|
|
132 (79%) |
36 (21%) |
68 (72%) |
27 (28%) |
Presence of Y-chromosome derivatives might be crucial for the health prognosis of X0 patients because of high risk of malignant gonadoblastoma development. Our results substantiate the following algorithm of mosaicism detection in patients with clinical features of TS (fig. 1). Molecular detection of Y chromosomes or its fragments with DNA probes for PCR reaction in DNA samples from peripheric blood should be taken as the 1st step. In the absense of Y-chromosome issue in blood, DNA analysis of the buccal mucosa cells might be suggested at the 2nd step. Positive results of DNA tests substantiate further thorough FISH analysis with Y-specific DNA-probes in lymphocytes and buccal mucosa cells for more precise determination of der(Y) structure at the 3rd step. However, not all patients with Y-chromosome derivatives reveal any masculinization traits. FISH-analysis of blood lymphocytes and buccal cells with X and Y-chromosome specific DNA probes should be recommended for all the rests of XO patients .
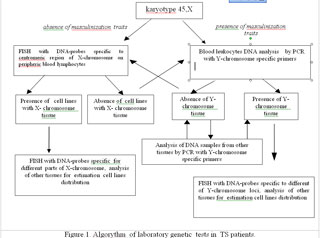
Figure.1. Algorythm of laboratory genetic tests in TS patients.
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

